The design of protein thermal stability is appealing for practical uses.
In a recent work [1] by Tian, Woodard, Whitney and Shakhnovich [see here] non-equilibrium Monte Carlo simulations were
effectively used to explore mutations of the Dihydrofolate Reductase
(DHFR) and their impact on both the stability and functionality of the
enzyme.
The key point of the work is the use of non-equilibrium Monte Carlo (MC) simulations. A protein is excited at different temperatures and its
"unfolding " as function of MC steps is recorded.
For each temperature, the average value of a given observable or order
parameter that describes the state of the protein, ie the root mean
square displacement with respect to the native state, the energy, the
gyration radius, depends on the simulation length -in the specific case
the number of MC steps. This relates to the fact that the transition
from the folded to the unfolded state, for a given temperature, is rate
limited by the free energy barrier dividing the two states. How this
dependence can be washed up when considering the effect of mutations?
The authors proved a nice recipe: first, a mutation affects the
thermodynamics of the system, formally the free energy difference
between folded and unfolded state, but also the kinetics for the
folded/unfolded transition, aka the free energy barrier dividing the two
state. It is possible to image that the thermodynamic effect is
mirrored on the change of free energy barrier via a scaling factor that measures
how the mutation influences the transition state of the folding/unfolding process.
Secondly, when considering the non-equilibrium MC simulations for both the
wild type and the mutant, the shift of the apparent melting temperature
(the temperature leading unfolding) of the mutated system with respect
to WT results independent from the simulation length. This can be formally showed, and the reader is invited to dig the work.
Using this strategy several mutations stabilizing the protein and that
maintain functionality were identified. I wonder whether this approach
can be used straightforwardly also for estimating the effect of
mutations on mechanical stability.
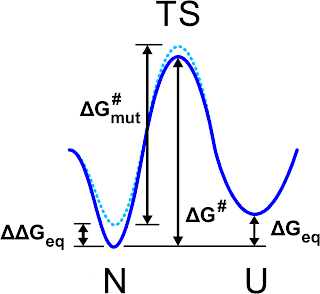 |
| Schematic view of the free energy profile for the folded (N) unfolded (U) states as it is pictured in Fig. 1 of Ref. 1 |
[1] J. Tian, JC Woodard, A. Whitney, EI Shakhnovich, Plos Comp Bio (2015) 11, e1004207.
No comments:
Post a Comment