 What factors contribute to protein stability at very extreme temperatures? What gain comes from entropy and enthalpy? And how to account for the delicate effect of temperature on molecular interactions like the hydrophobic and ionic ones? All this is tackled in a very intriguing work by Y. Matsura et al. "Thermodynamics of protein denaturation at temperature over 100°C: CutA1 mutant proteins substituted with hydrophobic and charged residues" recently published in Scientific Reports [see here]. By designing sequential mutations the authors were able to construct hyper-stable versions of the CutA1 protein and to extract the main thermodynamic parameters characterising their thermal stability. It is a very important work challenging both technical biochemical problems, like the aggregation of proteins generally occurring above 80°C, and the basic thermodynamics controlling protein stability over 100°C where for instance hydrophobic interactions cease to be entropically driven and ion-pairing can benefit from water dielectric constant decrease.
What factors contribute to protein stability at very extreme temperatures? What gain comes from entropy and enthalpy? And how to account for the delicate effect of temperature on molecular interactions like the hydrophobic and ionic ones? All this is tackled in a very intriguing work by Y. Matsura et al. "Thermodynamics of protein denaturation at temperature over 100°C: CutA1 mutant proteins substituted with hydrophobic and charged residues" recently published in Scientific Reports [see here]. By designing sequential mutations the authors were able to construct hyper-stable versions of the CutA1 protein and to extract the main thermodynamic parameters characterising their thermal stability. It is a very important work challenging both technical biochemical problems, like the aggregation of proteins generally occurring above 80°C, and the basic thermodynamics controlling protein stability over 100°C where for instance hydrophobic interactions cease to be entropically driven and ion-pairing can benefit from water dielectric constant decrease. Tuesday, November 17, 2015
Over 100°C
 What factors contribute to protein stability at very extreme temperatures? What gain comes from entropy and enthalpy? And how to account for the delicate effect of temperature on molecular interactions like the hydrophobic and ionic ones? All this is tackled in a very intriguing work by Y. Matsura et al. "Thermodynamics of protein denaturation at temperature over 100°C: CutA1 mutant proteins substituted with hydrophobic and charged residues" recently published in Scientific Reports [see here]. By designing sequential mutations the authors were able to construct hyper-stable versions of the CutA1 protein and to extract the main thermodynamic parameters characterising their thermal stability. It is a very important work challenging both technical biochemical problems, like the aggregation of proteins generally occurring above 80°C, and the basic thermodynamics controlling protein stability over 100°C where for instance hydrophobic interactions cease to be entropically driven and ion-pairing can benefit from water dielectric constant decrease.
What factors contribute to protein stability at very extreme temperatures? What gain comes from entropy and enthalpy? And how to account for the delicate effect of temperature on molecular interactions like the hydrophobic and ionic ones? All this is tackled in a very intriguing work by Y. Matsura et al. "Thermodynamics of protein denaturation at temperature over 100°C: CutA1 mutant proteins substituted with hydrophobic and charged residues" recently published in Scientific Reports [see here]. By designing sequential mutations the authors were able to construct hyper-stable versions of the CutA1 protein and to extract the main thermodynamic parameters characterising their thermal stability. It is a very important work challenging both technical biochemical problems, like the aggregation of proteins generally occurring above 80°C, and the basic thermodynamics controlling protein stability over 100°C where for instance hydrophobic interactions cease to be entropically driven and ion-pairing can benefit from water dielectric constant decrease. Sunday, November 1, 2015
Water helps life in extreme environments?
A few months ago we published an explorative work focusing on the possible contribution of water molecules buried in the interior of proteins to their different thermal stabilities [see here for the paper]. The study-case was a pair of homologous GTPase domains from a mesophilic and a hyperthermophilic organism, respectively. Now, we extended our approach by considering a large set of homologous pairs. Let's list the main findings. Firstly, for some homologues internal water gives a meaningful contribution to the stability gap in favour of the thermophilic variant. This was probed at ambient condition. Secondly, when considering the behaviour at high temperature, we found that thermophilic proteins are more keen to maintain their internal cavities wet, and therefore benefiting by this wetting. We propose that internal hydration can be viewed as an alternative tuneable variable for the engineering of proteins with enhanced stability. Enjoy the manuscript here.
Thursday, October 8, 2015
Fast and Cheap: the bioinformatics help to membrane protein thermal-stabilisation!
A very interesting work by Sauer et al. has been recently published in Biophysical J (here) and was highlighted by a nice comment by CG Tate (see here). What the story is about? Sauer and colleagues have deployed comparative bioinformatics tools to propose single point mutations for stabilising a membrane protein, the tetracycline antiporter from Bacillus subtilis (BsTetL). Out of the 22 mutations tested, 7 were proved to increase the thermal stability of the protein: almost 32% of success! This is a very high number. Why all this interest? Following the comment by Tate, membrane proteins -that are key targets for many drugs- are difficult to be crystallised. They must be isolated from the membrane environment and solubilised before crystallised. This is achieved by using detergents that protect the hydrophobic surface of the membrane protein from the contact with water. The stability of the protein/detergent complex is considered a pre-requisite for the success of the crystallisation process. While many effort has been placed to select/design the correct detergent to envelope the membrane protein, and alternative strategy consists in making the protein more stable. But select what to mutate, because of the highly complicate procedure, is essential. In this regards, the methodology proposed by Sauer et al. could be a breakthrough. Of course, questions remain. From the long research on thermophilic proteins we know that often stability trades functionality. Ergo, making the protein more stable could lock it in not functional states. This concerns is well explained in the Tate's comment. Go there, for an elegant discussion.
Thursday, September 24, 2015
In vivo stability! Cell type matters.....
In a very recent PNAS, J. Danielsson et al. [see here] present a very interesting work focusing on protein stability in cell. They used in-cell NMR to reconstruct the stability curve of the protein SOD1. The interesting finding is that when the protein is moved in two different types of cells, a bacterial (E. coli) and a mammalian cell (A2780) the protein is destabilised in both cases. Firstly, this finding questions the common believe that under crowding a protein gets stabilised because of an excluded volume effect. In short, if the available space is reduced because of the presence of a large numbers of macromolecules acting as crowders, the highly entropic and extended unfolded state should be unfavored. This picture is however very simplified since in both folded and unfolded states, a targeted protein interacts with its neighbours, and the results of these specific interactions, i.e. electrostatic, could alter the equilibrium favouring unfolding. The effect of different specific interactions, is actually probed by the authors, showing that by changing the local environment, moving from E.coli to a mammalian cell, the destabilisation effect is different. Last, but not least, the destabilisation results as an increase of the specific heat of unfolding that shrinks the stability curve. This calls for a particular effect of the crowders on the nature of the unfolded state. Stay tune, because the life of proteins in cell is where our interest is going....
| Molecular view of molecular crowding. Project at the Riken HPC center, Japan [see here] |
Monday, May 18, 2015
Designing thermal stability via non-equilibrium simulations
The design of protein thermal stability is appealing for practical uses.
In a recent work [1] by Tian, Woodard, Whitney and Shakhnovich [see here] non-equilibrium Monte Carlo simulations were
effectively used to explore mutations of the Dihydrofolate Reductase
(DHFR) and their impact on both the stability and functionality of the
enzyme.
The key point of the work is the use of non-equilibrium Monte Carlo (MC) simulations. A protein is excited at different temperatures and its
"unfolding " as function of MC steps is recorded.
For each temperature, the average value of a given observable or order
parameter that describes the state of the protein, ie the root mean
square displacement with respect to the native state, the energy, the
gyration radius, depends on the simulation length -in the specific case
the number of MC steps. This relates to the fact that the transition
from the folded to the unfolded state, for a given temperature, is rate
limited by the free energy barrier dividing the two states. How this
dependence can be washed up when considering the effect of mutations?
The authors proved a nice recipe: first, a mutation affects the
thermodynamics of the system, formally the free energy difference
between folded and unfolded state, but also the kinetics for the
folded/unfolded transition, aka the free energy barrier dividing the two
state. It is possible to image that the thermodynamic effect is
mirrored on the change of free energy barrier via a scaling factor that measures
how the mutation influences the transition state of the folding/unfolding process.
Secondly, when considering the non-equilibrium MC simulations for both the
wild type and the mutant, the shift of the apparent melting temperature
(the temperature leading unfolding) of the mutated system with respect
to WT results independent from the simulation length. This can be formally showed, and the reader is invited to dig the work.
Using this strategy several mutations stabilizing the protein and that
maintain functionality were identified. I wonder whether this approach
can be used straightforwardly also for estimating the effect of
mutations on mechanical stability.
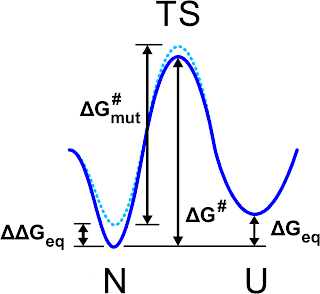 |
| Schematic view of the free energy profile for the folded (N) unfolded (U) states as it is pictured in Fig. 1 of Ref. 1 |
[1] J. Tian, JC Woodard, A. Whitney, EI Shakhnovich, Plos Comp Bio (2015) 11, e1004207.
Wednesday, May 13, 2015
A conserved structural element is a kinetic modulator of nucleotide exchange in the EF-Tu
A recent publication [1] reports that the P-loop, a conserved structural element in the catalytic domain of many different NTP-ases, might participate in modulating the nucleotide exchange rates for a broad class of NTP-ases.
The study was performed in detail on the EF-Tu protein, which catalyzes the GTP/GDP hydrolysis. Results show that the internal dynamics of the P-loop does not affect the nucleotide exchange rates, but rather P-loop forms a P-loop anchor via hydrogen bonds with another structural element of the protein, helix C. The P-loop anchor contributes to the activation entropy of the nucleotide exchange, and consequently modulates the nucleotide-binding kinetics.
Presently, two classes of P-anchors have been identified, depending on the nature of stabilization of the anchor, one class is hydrogen-bond stabilized, while the other is stabilized by the hydrophobic effect. The finding is consistent with the natural mechanism of the nucleotide exchange on the EF-Tu, where the nucleotide is exchanged upon the binding of another protein (EF-Ts) that disrupts the stability of the P-anchor, making the P-loop more flexible. The larger flexbility of the P-loop increases the entropic contribution to the activation free energy, resulting in faster nucleotide dissociation.
[1] Mercier E., Girodat D., Wieden H.-J., A conserved P-loop anchor limits the structural dynamics that mediate nucleotide dissociation in EF-Tu, JA - Sci. Rep., 2015.
Tuesday, May 5, 2015
And yet it functions!
A nice paper adressing the issue of how mesophilic/thermophilic enzymes
functions at differents temperatures is just out in Biochemistry [see
here].
The work by the group of EA Eisenmesser focuses on the behavior of the
cyclophilin enzyme from Geobacillus kaustophililus (GeoCyp), a bacterium
found in the the deep see sediment of the Mariana Trench, and compared
to its mesophilic homologous from humans (CypA). The study demonstrates
that, at variance with other mesophilic/thermophilic pair, here, the
thermophile maintains up to 70% of its catalytic power at low
temperature where most of thermophiles do not function or have very
limited activity. We have already posted on the "corresponding state
principle", introduced to explain why most of thermophiles lack activity
at ambient conditions. According to this view, the lack of activity is
due to the enhanced rigidity of the protein matrix which compromises
mobility essential to the catalytic turn-over. At the same time
mechanical rigidity is postulated as the source of the enhanced
stability of the protein and its resistance to thermal stress. The
universality of this principle has been questioned by showing that in
many cases thermophiles can be as flexible as their mesophilic variants
at the same thermodynamic conditions, thus stability is the results of a
smaller entropy penalty between folded (less entropic) and unfolded
(more entropic) states.
According to the work by Eisenmesser and coworkers, the thermophilic
GeoCyp is highly similar from the structural point of view to the human
CypA, and its dynamics at different timescales is comparable even if its
mobility seems more sensitive to temperature increases. What probably
causes the 30% drop of activity at low temperature (10°C) with respect
to activity at its optimal temperature (60°C) is a reduced local motion
of binding-site loop, which gating is affected by the presence of a
charged amino-acid, and a slightly less strong electric field measured
at the level of the catalytic site and supposed to ease the
isomerization of the peptide bond. In summary, this study shows us
another deviation from the common believe based on the observation of
reduced thermophilic activity at low temperatures. Nice work!
Friday, March 13, 2015
QM/MM study of Guanosine triphosphate (GTP) hydrolysis in homologous enzymes
Recently,
Swhwartz's group [1] reported their study on two bacterial enzymes
EcMTAN and VcMTAN, in addition to the inherent interest in the
function of these enzymes, they are also interesting test cases of how
very similiar enzymes with experimentally determined transition
states that are essentailly identical seem to have different reaction
mechanisms, since they are reported to have different binding
affinities for the same transition state analogue. So reaction
coordiante information is crucial to explain, and it is only
accessible from transition path sampling approaches, since all
experimental approaches report only average.
This
could also be the case for Guanosine triphosphate (GTP) hydrolysis in
their homologous enzymes. With different participation of differnt
enzyme environment, the GTP hydrolysis can happen in different ways,
even though the transition state are somehow analogue, and the
reaction mechanism in general is the same.
Phosphoryl
transfer reactions play a crucial role in widespread cellular
functions, ranging from the signaling [2] and stress-activated [3]
pathways to the biosynthesis of nucleic acids [4]. The key step of
action of dephosphorylating enzymes is the transfer of a phosporyl
group of GTP to acceptors such as water (hydolases), amino acids
residues (kinases) and other nucleotides (nucleoside monophosphate
kinases). Within the family of hydrolases, the low molecular weight
GTP-binding proteins (LMWGs) [5] hold a prominent position.
GTP + H2O → GDP + HPO4 =
One
member of this family is Cdc42/Cdc42GAP/GDP (PDB code 1GRN).
Different and conflicting proposals for the enzymatic mechanism have
been reported. They differ in three key points, which are also the
common points for GTP/GDP enzymatic reaction study:
(1) the formation of
the highly nucleophilic agent OH-; (2) its binding to GTP with the
formation of GDP and inorganic phosphate. (3) dissociative or
associative pathway in step (2)?
 Conclusion:
(1) H2O transfers its proton to Gln61, thus the highly nucleophilic
OH- is stabilized. Meantime, low-barrier hydrogen bond formed between
Lys16 and β-phosphate. (2) a chemical bond formed between
γ-phosphate and WAT oxygen. (3) the reactant is pre-built as a
dissociative reaction model.
Conclusion:
(1) H2O transfers its proton to Gln61, thus the highly nucleophilic
OH- is stabilized. Meantime, low-barrier hydrogen bond formed between
Lys16 and β-phosphate. (2) a chemical bond formed between
γ-phosphate and WAT oxygen. (3) the reactant is pre-built as a
dissociative reaction model.
Another
important member of this family is the Elongation factor Tu (EF-Tu).
There is a “star” amino acid that often under debates is the
histidine 85 (His85), who locates in the switch II region. One point
of view is that His85 acts as a base, it participates directly to the
OH- formation [6], the contrary is that His85 is not a base, but only
contribute to the stabilization of the transition state by hydrogen
binding [7]. However, it is proved that the replacement of His84 with
Ala reduces the rate constant of GTP hydrolysis more than 106
- fold. But not yet any fix conclusion of its role made.
There
is one interesting article from Nemukhin’s group [8] who talked
about two cases: His85 in (side chain of His85 approach the reaction
active site) and His85 out (side chain of His85 stays away from the
active site). The reactions’ transition and product states were
shown in Figure 2.
Conclusion: The character of TS is closer to the dissociative type
reaction mechanism than to the associative type. In the His85 in
case, His85 serves as a general base, while the His85 out case, the
reaction results as a consequence of proton transfer mediated by two
water molecules. The His85 in case show much lower activation barrier
which corresponds to experimental result. But it remains still a
interesting study point.
References:
[1] Matthew, I. Z.; Motley, M. W. ; Antoniou, D. ; Schramm
V. L . ; Schwartz S. D. J. Phys. Chem. B. 2015, 119,
3662-3668
[2] Takai,
Y.; Kishimoto, A.; Inoue, M.; Nishizuka, Y. J. Biol. Chem. 1977, 252,
7603-7609
[3]
Kyriakis, J. M.: Avruch, J. J. Biol. Chem. 1996, 271, 24313-24316
[4]
Koerner, J. F. Annu. Rev. Biochem. 1970, 39, 291-322.
[5]
Shinjo, K.; Koland, J. G.; Hart, M. J.; Narasimhan, V.; Johnson, D.
I.; Evans, T.; Cerione, R. A. Proc. Natl. Acad. Sci. U.S.A. 1990, 87,
9853-9857
[6]
Voorhees, M. R; Schmeing, T. M; Kelley, A. C; Ramakrishnan, K. V.
Science 2010, 330, 835
[7]
Daviter, T; Wieden, H; Rodnina, M. V. J. Mol. Biol. 2003, 332, 689.
[8]
Grigorenko, B.L.; Shadrina, M.S.; Topol I.A.; Collins J.R.; Nemukhin
A.V. Bichimica et Biophysica Acta 2008, 1784, 1908-1917
Wednesday, March 4, 2015
The lipids in archaea
Live in extreme conditions requires not only a set of stable proteins but also other resistant molecular components like membranes. In a recent work appeared in ScientificReports (1), see here, computer simulations have been used to explore the properties of a archaeal-like membranes that are formed by lipids chemically different with respect to the ones forming membranes in example in Bacteria. Lipids used by an archeon have hydrophobic tails linked together so that the molecule looks like triphasic with the following sequence glycerol head/hydrophobic tails/glycerol head. As consequence the membrane is assembled as a monolayer of lipids and not as a bilayer. On top of this, the chemical nature of the hydrophobic tail can be different as well. The main point of the manuscript is that because of the linked structure of the hydrophobic tails the archaeal-membrane is more dense and less permeable. This feature could be used for the optimal design of nano-structures representing resistant support in biotechnology.
[1] A.O. Chugnov et al. Scientific Reports, 2014, 4, 7462.
Tuesday, March 3, 2015
Protein Design
Extremophilic proteins represent a natural template to understand how enzymatic activity can be performed in non conventional conditions, i.e. high temperature, high salt concentration, low temperature, high pressure. Evolution has driven this migration of protein sequences/folds toward optimal states for different environments. How 'artificial evolution' can do the same job? In other words, how protein design can succeed in mutating proteins for changing their working milieu, or even incorporate new chemical activity? I propose to the readers two interesting reviews on this subject. One is from J.G. Saven group [1], the other from D. Baker and K.N Houk [2]. It is amazing how leading groups in the field have advanced the research combining multiple approaches. Enjoy!
[1] I. Samish, C.M. MacDermaid, J.M. Perez-Aguilar, J.G. Saven, Ann.Rev.Phys.Chem. (2011) 62, 129-149. (here)
[2] G. Kiss, N. Celebi-Olcum, R. Moretti, D. Baker, and K.N. Houk, Angew. Chem. Int. Ed. (2013) 52, 5700-5725. (here)
Subscribe to:
Comments (Atom)


